
Species distribution and antibiotic resistance in Shandong, China: 2010–2016
Introduction
Infectious diseases are most commonly found in the intensive care units (ICUs). Drug-resistant bacterial infections are the most challenging of these (1). With the widespread use of broad-spectrum antibiotics, the antibiotic susceptibilities and resistance patterns of infectious diseases might be changed accordingly (2). Optimal antibiotic use is crucial in septic patients (3). Unfortunately, 30% to 60% of the antibiotics used in the ICU were suboptimal, inappropriate and unnecessary (4-7). Continued monitoring of local epidemiological data is crucial to reflect current trends and appropriately guide disease management. The CHINET surveillance system has done lots of meaningful work to collect and report the changes and trends in antimicrobial resistance across China (8,9). However, data about the species distributions and antibiotic resistance of infections in local ICUs in East China are limited (10). Therefore, we aimed to establish a database of the species distributions and antibiotic resistance of infectious diseases in our local ICUs. We present this article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jeccm-20-121) (11).
Methods
Bacterial isolates and ethics
Clinical isolates from 34 ICUs participating in the Shandong Provincial antimicrobial resistance surveillance network were collected during 2010–2016. All of the pathogens isolated were included in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review boards of Qianfoshan Hospital, Shandong University (ethics approval number: 2009-S068). The institutional review board specifically approved the informed consent waiver because of the anonymous and purely observational nature of this study.
Antimicrobial susceptibility testing
The susceptibilities were determined by the disk diffusion method in accordance with the Clinical and Laboratory Standards Institute guidelines (12,13).
Data acquisition
Data collected from 34 ICUs were recorded by WHONET 5.6 software (14,15) (http://www.whonet.org). A database was established in WHONET 5.6 for samples from the same hospital with all of the antibiotics used in the antimicrobial susceptibility test selected from the list of antibiotics. Patient information, the sample category, and the pathogen index according to the rule of WHONET were stored while documenting the pathogen information. Double entry and validation were performed. Any missing data were retrieved from the hospital medical system. At each participating hospital, databases were updated quarterly, and data from all 34 hospitals were compiled annually.
Analysis
ICUs participating in the Shandong Provincial antimicrobial resistance surveillance network held an annual meeting. The ICU department of Qianfoshan Hospital collected the WHONET databases from the 34 hospitals, which were shared and discussed during the annual meeting. Figure S1 shows the location of the 34 ICUs in Shandong Province.
Statistics
Numbers and percentages were used to report the distributions of specimens. Chi-square tests were performed to examine the statistical differences of the proportions of different isolates in difference specimens. The trend Chi-square tests were performed for the changes of antibiotic resistance rates through the seven years. The statistical tests were performed using SAS 9.4.
Results
Distribution of isolates
A total of 61,901 clinical isolates of pathogens, from 61,564 samples, were collected, including sputum samples (76.8%, n=47,281), blood cultures (11.3%, n=6,956), urine samples (7.5%, n=4,617), pyogenic fluids (1.6%, n=985), venous catheter (1.5%, n=923) and cerebrospinal fluids (1.0%, n=615). The ratio of samples had no significant variation during 2010–2016. The 61,901 pathogens included Gram-negative bacteria (80.1%, n=49,583), Gram-positive bacteria (13.4%, n=8,289) and fungi (6.5%, n=4,029).
The most common isolations in different specimens were reported in Table 1. The isolation distributions in each kind of specimen were significantly different (P<0.001).
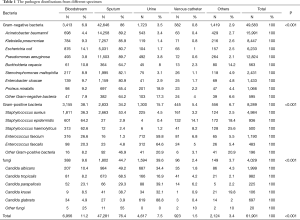
Full table
The 49,583 Gram-negative bacteria included Acinetobacter baumannii (32.3%, n=15,991), Pseudomonas aeruginosa (25.9%, n=12,824), Klebsiella pneumoniae (17.0%, n=8,447), Escherichia coli (12.6%, n=6,233), Stenotrophomonas maltophilia (4.9%, n=2,431), Enterobacter cloacae (2.9%, n=1,433), Proteus mirabilis (2.2%, n=1,066), Burkholderia mirabilis (1.1%, n=563) and others (1.1%, n=595).
The 8,289 Gram-positive bacteria included Staphylococcus aureus (60.1%, n=4,984), Enterococcus aureus (20.2%, n=1,673), Staphylococcus epidermidis (11.3%, n=936), Staphylococcus haemolyticus (6.0%, n=500) and others (2.4%, n=196). The 2033 Enterococcus aureus bacteria included Enterococcus faecium (71.1%, n=1,190) and Enterococcus faecalis (28.9%, n=483).
The 4,029 fungi included Aspergillus species (0.5%, n=20) and Candida species (99.5%, n=4,009). The 4,009 Candida species included Candida albicans (49.9%, n=1,999) and non-albicans (50.1%, n=2,010). The 2010 non-albicans Candida species included Candida tropicalis (48.9%, n=982), Candida glabrata (34.6%, n=697), Candida parapsilosis (11.2%, n=225) and Candida krusei (5.3%, n=106).
Antimicrobial susceptibility testing
In Enterobacteriaceae bacteria, the resistance of Klebsiella pneumoniae to Imipenem and Cefoperazone/sulbactam increased (P<0.001) to Piperacillin/tazobactam, Amikacin, Ciprofloxacin, Levofloxacin, Ceftazidime, Cefepime and Ceftriaxone decreased (P<0.001). The resistance of Escherichia coli to Imipenem, Meropenem, Cefoperazone/sulbactam, Amikacin, Ceftazidime, Cefepime, Levofloxacin, Ciprofloxacin and Ceftriaxone increased (P<0.001); to Piperacillin/tazobactam decreased (P<0.001). The resistance of Proteus mirabilis to Cefoperazone/sulbactam increased; to Meropenem, Piperacillin/tazobactam, Amikacin, Ceftazidime, Cefepime (P<0.001), Levofloxacin, Ciprofloxacin and Ceftriaxone (0.42, 0.44, 0.33, respectively) decreased. The resistance of Enterobacter cloacae to Meropenem, Piperacillin/tazobactam, Cefoperazone/sulbactam, Ciprofloxacin increased (P<0.05); to Amikacin, Cefepime, Levofloxacin, Ceftazidime and Ceftriaxone (P<0.001) decreased. The Enterobacteriaceae bacteria kept a high level of sensitivity to Carbapenems, Piperacillin-tazobactam, Cefoperazone-sulbactam and Amikacin (Figure 1 and Table 2).
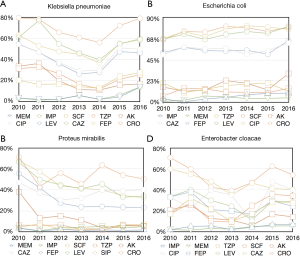
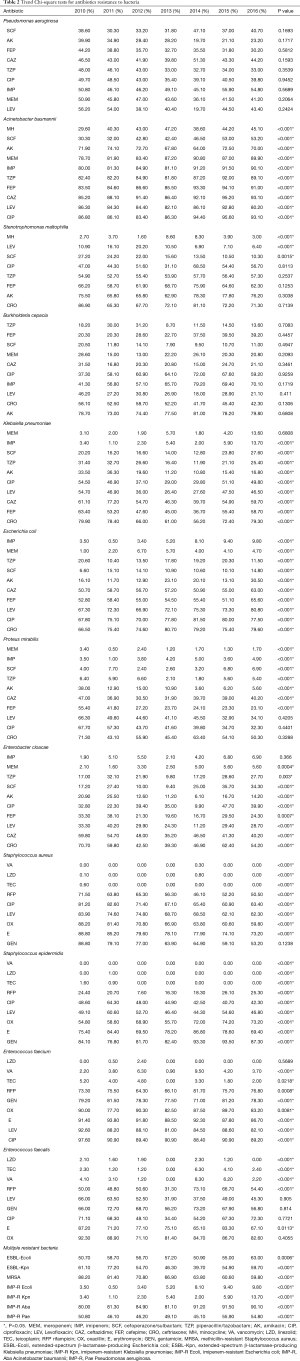
Full table
In non-fermentative Gram-negative bacteria, the resistance of Acinetobacter baumannii to Minocycline, Cefoperazone/sulbactam, Meropenem, Imipenem, Piperacillin/tazobactam, Cefepime, Ceftazidime, Levofloxacin and Ciprofloxacin increased (P<0.001); to Amikacin and Levofloxacin decreased (P<0.001). The resistance of Stenotrophomonas maltophilia to Minocycline increased (P<0.001); to Levofloxacin, Cefoperazone/sulbactam decreased (P<0.001, p=0.0015, respectively). There were no trend identified of Pseudomonas aeruginosa and Burkholderia cepacia (Figure 2 and Table 2).
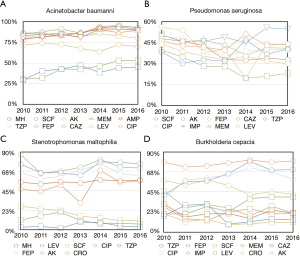
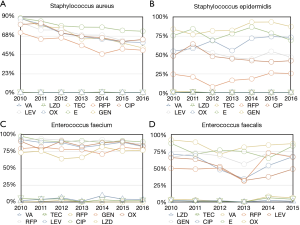
In Gram-positive bacteria, the resistance of Staphylococcus aureus to Rifampicin, Ciprofloxacin, Levofloxacin, Oxacillin, Erythromycin (P<0.001) decreased. The resistance of Staphylococcus epidermidis to Rifampicin, Oxacillin and Gentamicin (P<0.001) increased; to Ciprofloxacin, Levofloxacin and Erythromycin (P<0.001) decreased. The resistance of Enterococcus faecium to Vancomycin, Rifampicin (P<0.001) increased; to Teicoplanin, Gentamicin, Oxacillin, Erythromycin, Levofloxacin and Ciprofloxacin (P<0.05) decreased. The resistance of Enterococcus faecalis to Teicoplanin, Rifampicin (P<0.001) increased; to Linezolid, Vancomycin and Erythromycin (P<0.05) decreased (Figure 3 and Table 2).
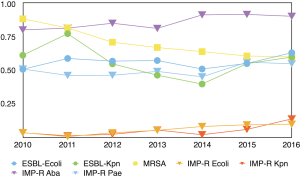
For the most common and concerning multiple-drug-resistant bacteria, extended-spectrum β-lactamase-producing Escherichia coli, Imipenem-resistant Klebsiella pneumoniae, Imipenem-resistant Escherichia coli, Imipenem-resistant Klebsiella pneumoniae, and Imipenem-resistant Acinetobacter baumannii and Imipenem-resistant Pseudomonas aeruginosa (P<0.001) increased. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae, Methicillin-resistant Staphylococcus aureus (P<0.001) decreased (Figure 4 and Table 2).
Discussion
Our research indicated that non-fermentative bacteria are the most commonly isolated pathogens in our ICUs, representing more than 60% of the overall isolates, in comparison with 33% in ICUs in Taiwan (16). The high proportion of non-fermentative bacteria isolated might be related to the following causes. First, we conducted frequent routine bacterial cultures. The isolated non-fermentative bacteria were, to some extent, colonization rather than pathogenic bacteria, which might limit the meaning of their prevalence. Also, it should be noted that frequent cultures from the same patients might lead to the same isolates being included in the data collection over and over again. Second, we might have a suboptimal level of infection control. Since non-fermentative bacteria widely exist in the environment, suboptimal hand hygiene might result in the transmission of this type of bacteria (17,18). Our results prompted us to improve the hand hygiene and project-related infection control schemes in our ICUs.
From these data, the overall level of resistance to Carbapenems was on the rise. Notably, the resistance of Acinetobacter baumannii increased tremendously and rapidly. We noticed that the carbapenems resistance Klebsiella pneumoniae decreased in the first several years and then gradually increased in recent years. We compared our results with the CHINET surveillance (www.chinets.com). The fluctuations of the carbapenems resistant Klebsiella pneumonia rates through the seven years were similar with our results in Beijing. However, the specific rates differed tremendously from regions across China. Carbapenem resistance is a tough problem worldwide (19-21). There is a lack of potent and effective antimicrobials to Carbapenem-resistant Gram-negative bacterial infections since such strains are usually extensively drug resistant (22). As novel potent antibiotics could not be counted on, the prevention of clonal dissemination and the avoidance of Carbapenem overuse are vital to preventing the spread of these bacteria (23). Surveillance of bacteria distribution and drug resistance is also important (24).
There are strengths in our study. We conducted a continued, long-period surveillance in our local ICUs and included almost all of the major cities in Shandong Province. The data should be more related to our local clinical conditions in comparison with the surveillances of various medical departments and areas across China. To sum up, we shared the data and held a meeting annually between all member ICUs. Therefore, we kept adequate communication and made efforts to reduce the variations in bacterial cultures, data processing and also infection controls. There are several limitations to our study. First and foremost, this study did not make a distinction between pathogenic and colonized bacteria. For the vast majority of candida in sputum, the isolates had no much clinical meaning. Besides, we did not further classify the sources of specific sputum. We reported the isolates; yet, the relations between the isolates and infections were not analyzed in this study. Moreover, the clinical severity data of these bacteria were not recorded; thus the associations between bacteria and clinical severity were uncertain. Second, there was no controlled design in this study, so the relations between the variations of bacterial resistance and clinical outcomes were unclear. And without clinical interventions recorded, we could not analyze the associations of resistance changes and clinical practice. Third, the data were recorded according to the reports from the microbiology laboratories and no further genetic testing were conducted. The further identification of resistant strains was therefore limited. Fourth, we described the distribution and resistance patterns of pathogens in ICUs in our region. However, resistance patterns vary in different regions. The meaning for other regions is limited.
Conclusions
The surveillance showed that Gram-negative bacteria, especially non-fermentative Gram-negative strains, are the most commonly isolated in the ICUs in Shandong, China. Acinetobacter baumannii and Pseudomonas aeruginosa are the most commonly isolated bacteria. Non-fermentative Gram-negative bacteria had the highest antimicrobial resistance rates. Acinetobacter baumannii had a significantly higher level of resistance to antimicrobial agents than Pseudomonas aeruginosa. Escherichia coli showed higher antimicrobial resistance than Klebsiella pneumoniae. Enterococcus faecium showed more resistance toward all antibiotics than Enterococcus faecalis. extended-spectrum β-lactamase-producing Escherichia coli, Imipenem-resistant Klebsiella pneumoniae, Imipenem-resistant Escherichia coli, Imipenem-resistant Klebsiella pneumoniae, and Imipenem-resistant Acinetobacter baumannii and Imipenem-resistant Pseudomonas aeruginosa increased in our region.
Acknowledgments
The abstract of this work was reported on the 41th Annual Conference on Shock (Arizona), and published on Shock 2018;49(6s):36. Here are acknowledgements to the participating ICUs of the following hospitals: Shandong Qianfoshan Hospital, Qilu Hospital of Shandong University, General Hospital of Jinan Command, The Second Affiliated Hospital of Shandong University, Shandong Chest Hospital, East District of Shandong Provincial Hospital of Traditional Chinese Medicine, West District of Shandong Provincial Hospital of Traditional Chinese Medicine, Jinan City Central Hospital, The Forth Jinan Municipal Hospital, Affiliated Hospital of Weifang Medical College, Weifang People’s Hospital, Affiliated Hospital of Binzhou Medical College, Binzhou People’s Hospital, Dezhou People’s Hospital, General Hospital of Shengli Oil Field in Dongying, Dongying People’s Hospital, Heze Municiple Hospital, Jining First People’s Hospital, Affiliated Hospital of Jining Medical College, Qingdao Navy 401 Hospital, Affiliated Hospital of Qiingdao University Medical College, East District of Affiliated Hospital of Qiingdao University Medical College, East District of Oingdao Municipal Hospital, Qingdao Haici Hospital, Yuhuangding Hospital, Yantaishan Mountain Hospital, Liaocheng People’s Hospital, Rizhao People’s Hospital, Affiliated Hospital of Taishan Medical College, Central Hospital, Tengzhou Central People’s Hospital, Weihai Municipal Hospital, Wendeng Central Hospital, Zibo Central Hospital. The authors want to thank Yang Wang, Qi Zhang and Ge Chen for the consultant and help in statistics.
Funding: The research was supported by Shandong Provincial Natural Science Foundation (ZR2015CL012), and grants from the Medical Science and Technology Development Program of Shandong Province (2017WS363 and 2017WS761).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jeccm-20-121
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jeccm-20-121
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jeccm-20-121). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review boards of Qianfoshan Hospital, Shandong University (ethics approval number: 2009-S068). The institutional review board specifically approved the informed consent waiver because of the anonymous and purely observational nature of this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arnold HM, Micek ST, Skrupky LP, et al. Antibiotic stewardship in the intensive care unit. Semin Respir Crit Care Med 2011;32:215-27. [Crossref] [PubMed]
- Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance - the need for global solutions. Lancet Infect Dis 2013;13:1057-98. [Crossref] [PubMed]
- Luyt CE, Bréchot N, Trouillet JL, et al. Antibiotic stewardship in the intensive care unit. Crit Care 2014;18:480. [Crossref] [PubMed]
- Bergmans DC, Bonten MJ, Gaillard CA, et al. Indications for antibiotic use in ICU patients: a one-year prospective surveillance. J Antimicrob Chemother 1997;39:527-35. [Crossref] [PubMed]
- Roberts JA, Paul SK, Akova M, et al. DALI: defining antibiotic levels in intensive care unit patients: are current beta-lactam antibiotic doses sufficient for critically ill patients? Clin Infect Dis 2014;58:1072-83. [Crossref] [PubMed]
- Kollef MH. Optimizing antibiotic therapy in the intensive care unit setting. Crit Care 2001;5:189-95. [Crossref] [PubMed]
- Kollef MH, Fraser VJ. Antibiotic resistance in the intensive care unit. Ann Intern Med 2001;134:298-314. [Crossref] [PubMed]
- Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect 2016;22 Suppl 1:S9-14. [Crossref] [PubMed]
- Hu F, Zhu D, Wang F, et al. CHINET 2014 surveillance of bacterial resistance in China. Chin J Infect Chemother 2015;15:401-10.
- Zhang Y, Yao Z, Zhan S, et al. Disease burden of intensive care unit-acquired pneumonia in China: a systematic review and meta-analysis. Int J Infect Dis 2014;29:84-90. [Crossref] [PubMed]
- von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: guidelines for reporting observational studies. Int J Surg 2014;12:1495-9. [Crossref] [PubMed]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Eighteenth informational supplement document. M100-S18. Wayne, PA: CLSI, 2006.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twentieth Informational Supplement Document 2010;30:108-14.
- Park R, O'Brien TF, Huang SS, et al. Statistical detection of geographic clusters of resistant Escherichia coli in a regional network with WHONET and SaTScan. Expert Rev Anti Infect Ther 2016;14:1097-107. [Crossref] [PubMed]
- Manual on antimicrobial resistance and susceptibility testing. Division of emerging and other communicable diseases surveillance and control. WHO antimicrobial resistance monitoring programme. Geneva: WHO, 1997.
- Liu KS, Wang YT, Lai YC, et al. Antimicrobial resistance of bacterial isolates from respiratory care wards in Taiwan: a horizontal surveillance study comparison of the characteristics of nosocomial infection and antimicrobial-resistant bacteria in adult Intensive Care Units and two respiratory care facilities for mechanically ventilated patients at a tertiary care centre in Taiwan. Int J Antimicrob Agents 2011;37:10-5. [Crossref] [PubMed]
- Fonguh S, Uwineza A, Catry B, et al. Belgian hand hygiene campaigns in ICU, 2005-2015. Arch Public Health 2016;74:47. [Crossref] [PubMed]
- Huang HP, Chen B, Wang HY, et al. The efficacy of daily chlorhexidine bathing for preventing healthcare-associated infections in adult intensive care units. Korean J Intern Med 2016;31:1159-70. [Crossref] [PubMed]
- European Centre for Disease Prevention and Control. Annual epidemiological report. Antimicrobial resistance and healthcare-associated infections, 2014. Stockholm: ECDC, 2014.
- Hu F, Chen S, Xu X, et al. Emergence of carbapenem-resistant Enterobacteriaceae clinical isolates from a teaching hospital in Shanghai, China. J Med Microbiol 2012;61:132-6. [Crossref] [PubMed]
- Chen S, Hu F, Liu Y, et al. Detection and spread of carbapenem-resistant Citrobacter freundii in a teaching hospital in China. Am J Infect Control 2011;39:e55-60. [Crossref] [PubMed]
- Centers for Disease Control and Prevention (CDC). Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep 2009;58:256-60. [PubMed]
- Borer A, Saidel-Odes L, Riesenberg K, et al. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol 2009;30:972-6. [Crossref] [PubMed]
- Qin X, Yang Y, Hu F, et al. Hospital clonal dissemination of Enterobacter aerogenes producing carbapenemase KPC-2 in a Chinese teaching hospital. J Med Microbiol 2014;63:222-8. [Crossref] [PubMed]
Cite this article as: Jiang Z, Zhang Y, Zhu Y, Zhang L, Li C, Xie J, Yao Y, Du B, Xi X, Qu Y. Species distribution and antibiotic resistance in Shandong, China: 2010–2016. J Emerg Crit Care Med 2021;5:5.


